|
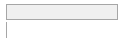
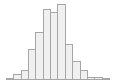
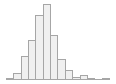
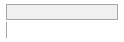
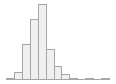
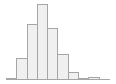
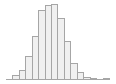
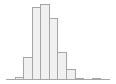
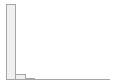
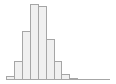
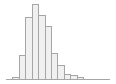
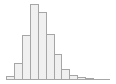
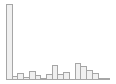
specimen_results_read
[POSIXct, POSIXt]
|
|
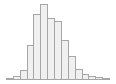
min : 2015-05-20 14:00:07
|
|
med : 2015-08-03 09:05:48.5
|
|
max : 2016-05-31 15:29:01
|
|
range : 1y 0m 11d 1H 28M 54S
|
|
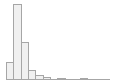
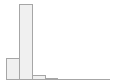
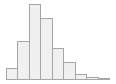
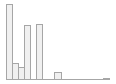
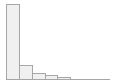
4910 distinct values
|
|
497053 (96.4%)
|
|
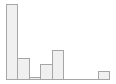
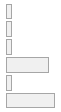
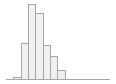
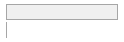
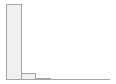
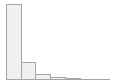
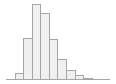
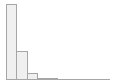
lipaemic [factor]
|
- Not
|
- Severity 1 (on a scale of
|
- Severity 2 (on a scale of
|
- Severity 3 (on a scale of
|
- Severity 4 (on a scale of
|
- Severity 5 (on a scale of
|
- Severity 6 (on a scale of
|
|
|
14740
|
(
|
80.3%
|
)
|
|
2595
|
(
|
14.1%
|
)
|
|
698
|
(
|
3.8%
|
)
|
|
159
|
(
|
0.9%
|
)
|
|
114
|
(
|
0.6%
|
)
|
|
10
|
(
|
0.1%
|
)
|
|
44
|
(
|
0.2%
|
)
|
|
|
497053 (96.4%)
|
|
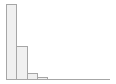
icteric [factor]
|
- Not
|
- Severity 1 (on a scale of
|
- Severity 2 (on a scale of
|
- Severity 3 (on a scale of
|
- Severity 4 (on a scale of
|
- Severity 5 (on a scale of
|
- Severity 6 (on a scale of
|
|
|
18298
|
(
|
99.7%
|
)
|
|
14
|
(
|
0.1%
|
)
|
|
3
|
(
|
0.0%
|
)
|
|
1
|
(
|
0.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
44
|
(
|
0.2%
|
)
|
|

|
497053 (96.4%)
|
|
haemolysed
[factor]
|
- Not
|
- Severity 1 (on a scale of
|
- Severity 2 (on a scale of
|
- Severity 3 (on a scale of
|
- Severity 4 (on a scale of
|
- Severity 5 (on a scale of
|
- Severity 6 (on a scale of
|
|
|
18108
|
(
|
98.6%
|
)
|
|
177
|
(
|
1.0%
|
)
|
|
26
|
(
|
0.1%
|
)
|
|
3
|
(
|
0.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
44
|
(
|
0.2%
|
)
|
|

|
497053 (96.4%)
|
|
turbid [factor]
|
|
|
18350
|
(
|
99.9%
|
)
|
|
10
|
(
|
0.1%
|
)
|
|
|
497053 (96.4%)
|
|
sample_note
[character]
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 16 others ]
|
|
|
42
|
(
|
22.5%
|
)
|
|
30
|
(
|
16.0%
|
)
|
|
29
|
(
|
15.5%
|
)
|
|
21
|
(
|
11.2%
|
)
|
|
11
|
(
|
5.9%
|
)
|
|
10
|
(
|
5.3%
|
)
|
|
9
|
(
|
4.8%
|
)
|
|
7
|
(
|
3.7%
|
)
|
|
4
|
(
|
2.1%
|
)
|
|
4
|
(
|
2.1%
|
)
|
|
20
|
(
|
10.7%
|
)
|
|
|
515226 (100.0%)
|
|
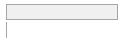
alb_gl [numeric]
|
|
Mean (sd) : 42.1 (2.9)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 42.2 ≤ 54.8
|
|
IQR (CV) : 3.5 (0.1)
|
|
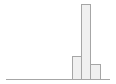
1581 distinct values
|
|
498393 (96.7%)
|
|
alb_invalid
[factor]
|
|
|
17018
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
498393 (96.7%)
|
|
alb_below_range
[factor]
|
|
|
17020
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
498393 (96.7%)
|
|
alb_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 8 others ]
|
|
|
47
|
(
|
44.3%
|
)
|
|
27
|
(
|
25.5%
|
)
|
|
8
|
(
|
7.5%
|
)
|
|
6
|
(
|
5.7%
|
)
|
|
4
|
(
|
3.8%
|
)
|
|
2
|
(
|
1.9%
|
)
|
|
1
|
(
|
0.9%
|
)
|
|
1
|
(
|
0.9%
|
)
|
|
1
|
(
|
0.9%
|
)
|
|
1
|
(
|
0.9%
|
)
|
|
8
|
(
|
7.5%
|
)
|
|

|
515307 (100.0%)
|
|
alb_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
17020
|
(
|
100.0%
|
)
|
|

|
498393 (96.7%)
|
|
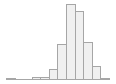
alt_ul [numeric]
|
|
Mean (sd) : 22.1 (17)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 17.7 ≤ 535.5
|
|
IQR (CV) : 12.2 (0.8)
|
|
898 distinct values
|
|
497230 (96.5%)
|
|
alt_invalid
[factor]
|
|
|
18171
|
(
|
99.9%
|
)
|
|
12
|
(
|
0.1%
|
)
|
|
|
497230 (96.5%)
|
|
alt_below_range
[factor]
|
|
|
18179
|
(
|
100.0%
|
)
|
|
4
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
alt_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result is
|
|
[ 57 others ]
|
|
|
530
|
(
|
76.0%
|
)
|
|
42
|
(
|
6.0%
|
)
|
|
32
|
(
|
4.6%
|
)
|
|
13
|
(
|
1.9%
|
)
|
|
7
|
(
|
1.0%
|
)
|
|
5
|
(
|
0.7%
|
)
|
|
3
|
(
|
0.4%
|
)
|
|
3
|
(
|
0.4%
|
)
|
|
3
|
(
|
0.4%
|
)
|
|
2
|
(
|
0.3%
|
)
|
|
57
|
(
|
8.2%
|
)
|
|

|
514716 (99.9%)
|
|
alt_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
18183
|
(
|
100.0%
|
)
|
|

|
497230 (96.5%)
|
|
apoa1_mgdl
[numeric]
|
|
Mean (sd) : 133.7 (23.3)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 131.6 ≤ 292.7
|
|
IQR (CV) : 28.5 (0.2)
|
|
1306 distinct values
|
|
497230 (96.5%)
|
|
apoa1_invalid
[factor]
|
|
|
18138
|
(
|
99.8%
|
)
|
|
45
|
(
|
0.2%
|
)
|
|
|
497230 (96.5%)
|
|
apoa1_below_range
[factor]
|
|
|
18183
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
apoa1_note
[character]
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- “Result obtained from 1 i
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {LIH test n
|
- AU680 analysis: {HEMOL =
|
- “Result obtained from 1 i
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 11 others ]
|
|
|
29
|
(
|
46.0%
|
)
|
|
7
|
(
|
11.1%
|
)
|
|
4
|
(
|
6.3%
|
)
|
|
3
|
(
|
4.8%
|
)
|
|
3
|
(
|
4.8%
|
)
|
|
2
|
(
|
3.2%
|
)
|
|
1
|
(
|
1.6%
|
)
|
|
1
|
(
|
1.6%
|
)
|
|
1
|
(
|
1.6%
|
)
|
|
1
|
(
|
1.6%
|
)
|
|
11
|
(
|
17.5%
|
)
|
|

|
515350 (100.0%)
|
|
apoa1_machine_id
[integer]
|
|
Min : -1019429
|
|
Mean : -1019349.6
|
|
Max : -1019349
|
|
|
-1019429
|
:
|
146
|
(
|
0.8%
|
)
|
|
-1019349
|
:
|
18037
|
(
|
99.2%
|
)
|
|

|
497230 (96.5%)
|
|
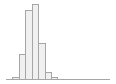
apob_mgdl
[numeric]
|
|
Mean (sd) : 83.8 (21.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 83 ≤ 311
|
|
IQR (CV) : 28.3 (0.3)
|
|
1221 distinct values
|
|
497230 (96.5%)
|
|
apob_invalid
[factor]
|
|
|
18181
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
apob_below_range
[factor]
|
|
|
18183
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
apob_note
[character]
|
- AU680 result: {Result may
|
- AU680 result: {Result is
|
- AU680 result: {Result may
|
- AU680 result: {LIH test n
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result is
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 46 others ]
|
|
|
242
|
(
|
42.8%
|
)
|
|
164
|
(
|
29.0%
|
)
|
|
28
|
(
|
5.0%
|
)
|
|
26
|
(
|
4.6%
|
)
|
|
21
|
(
|
3.7%
|
)
|
|
16
|
(
|
2.8%
|
)
|
|
5
|
(
|
0.9%
|
)
|
|
4
|
(
|
0.7%
|
)
|
|
4
|
(
|
0.7%
|
)
|
|
4
|
(
|
0.7%
|
)
|
|
51
|
(
|
9.0%
|
)
|
|

|
514848 (99.9%)
|
|
apob_machine_id
[integer]
|
|
Min : -1019429
|
|
Mean : -1019350.8
|
|
Max : -1019349
|
|
|
-1019429
|
:
|
403
|
(
|
2.2%
|
)
|
|
-1019349
|
:
|
17780
|
(
|
97.8%
|
)
|
|

|
497230 (96.5%)
|
|
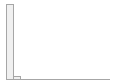
ast_ul [numeric]
|
|
Mean (sd) : 28.3 (17.8)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 24.7 ≤ 730.1
|
|
IQR (CV) : 10.4 (0.6)
|
|
860 distinct values
|
|
497230 (96.5%)
|
|
ast_invalid
[factor]
|
|
|
18175
|
(
|
100.0%
|
)
|
|
8
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
ast_below_range
[factor]
|
|
|
18178
|
(
|
100.0%
|
)
|
|
5
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
ast_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 95 others ]
|
|
|
291
|
(
|
48.1%
|
)
|
|
148
|
(
|
24.5%
|
)
|
|
32
|
(
|
5.3%
|
)
|
|
15
|
(
|
2.5%
|
)
|
|
5
|
(
|
0.8%
|
)
|
|
4
|
(
|
0.7%
|
)
|
|
3
|
(
|
0.5%
|
)
|
|
3
|
(
|
0.5%
|
)
|
|
3
|
(
|
0.5%
|
)
|
|
3
|
(
|
0.5%
|
)
|
|
98
|
(
|
16.2%
|
)
|
|

|
514808 (99.9%)
|
|
ast_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
18183
|
(
|
100.0%
|
)
|
|

|
497230 (96.5%)
|
|
chol_mmoll
[numeric]
|
|
Mean (sd) : 4.7 (1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 4.6 ≤ 16.2
|
|
IQR (CV) : 1.2 (0.2)
|
|
655 distinct values
|
|
497230 (96.5%)
|
|
chol_invalid
[factor]
|
|
|
18181
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
chol_below_range
[factor]
|
|
|
18183
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
chol_note
[character]
|
- AU680 result: {Result may
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 11 others ]
|
|
|
29
|
(
|
34.9%
|
)
|
|
22
|
(
|
26.5%
|
)
|
|
8
|
(
|
9.6%
|
)
|
|
4
|
(
|
4.8%
|
)
|
|
3
|
(
|
3.6%
|
)
|
|
2
|
(
|
2.4%
|
)
|
|
1
|
(
|
1.2%
|
)
|
|
1
|
(
|
1.2%
|
)
|
|
1
|
(
|
1.2%
|
)
|
|
1
|
(
|
1.2%
|
)
|
|
11
|
(
|
13.3%
|
)
|
|

|
515330 (100.0%)
|
|
chol_machine_id
[integer]
|
|
Min : -1019429
|
|
Mean : -1019349.3
|
|
Max : -1019349
|
|
|
-1019429
|
:
|
66
|
(
|
0.4%
|
)
|
|
-1019349
|
:
|
18117
|
(
|
99.6%
|
)
|
|

|
497230 (96.5%)
|
|
crea_umoll
[numeric]
|
|
Mean (sd) : 65.3 (26.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 62 ≤ 1536
|
|
IQR (CV) : 20 (0.4)
|
|
207 distinct values
|

|
497230 (96.5%)
|
|
crea_invalid
[factor]
|
|
|
18181
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
crea_below_range
[factor]
|
|
|
18175
|
(
|
100.0%
|
)
|
|
8
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
crea_note
[character]
|
- AU680 result: {Result is
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 result: {Result is
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {OD is high
|
- “Result confirmed by reru
|
|
[ 26 others ]
|
|
|
54
|
(
|
32.3%
|
)
|
|
36
|
(
|
21.6%
|
)
|
|
26
|
(
|
15.6%
|
)
|
|
8
|
(
|
4.8%
|
)
|
|
6
|
(
|
3.6%
|
)
|
|
4
|
(
|
2.4%
|
)
|
|
2
|
(
|
1.2%
|
)
|
|
2
|
(
|
1.2%
|
)
|
|
2
|
(
|
1.2%
|
)
|
|
1
|
(
|
0.6%
|
)
|
|
26
|
(
|
15.6%
|
)
|
|

|
515246 (100.0%)
|
|
crea_machine_id
[integer]
|
|
Min : -1019429
|
|
Mean : -1019349
|
|
Max : -1019349
|
|
|
-1019429
|
:
|
2
|
(
|
0.0%
|
)
|
|
-1019349
|
:
|
18181
|
(
|
100.0%
|
)
|
|

|
497230 (96.5%)
|
|
cysc_mgl [numeric]
|
|
Mean (sd) : 0.9 (0.3)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.8 ≤ 7.7
|
|
IQR (CV) : 0.2 (0.3)
|
|
689 distinct values
|
|
498930 (96.8%)
|
|
cysc_invalid
[factor]
|
|
|
16481
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
498930 (96.8%)
|
|
cysc_below_range
[factor]
|
|
|
16483
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
498930 (96.8%)
|
|
cysc_note
[character]
|
- “Result checked 0.41”
|
- “Result confirmed by repe
|
- “Result confirmed by repe
|
|
|
1
|
(
|
33.3%
|
)
|
|
1
|
(
|
33.3%
|
)
|
|
1
|
(
|
33.3%
|
)
|
|

|
515410 (100.0%)
|
|
cysc_machine_id
[integer]
|
|
Min : -1003889
|
|
Mean : -1003887.9
|
|
Max : -1003887
|
|
|
-1003889
|
:
|
7815
|
(
|
47.4%
|
)
|
|
-1003887
|
:
|
8668
|
(
|
52.6%
|
)
|
|

|
498930 (96.8%)
|
|
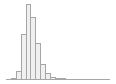
fib_gl [numeric]
|
|
Mean (sd) : 3.1 (0.8)
|
|
min ≤ med ≤ max:
|
|
0.9 ≤ 3 ≤ 10.7
|
|
IQR (CV) : 0.9 (0.2)
|
|
474 distinct values
|
|
506028 (98.2%)
|
|
fib_invalid
[factor]
|
|
|
9385
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
506028 (98.2%)
|
|
fib_below_range
[factor]
|
|
|
9385
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
506028 (98.2%)
|
|
fib_note
[character]
|
- Nephelometer result: { “P
|
|

|
515412 (100.0%)
|
|
fib_machine_id
[integer]
|
|
Min : -1003889
|
|
Mean : -1003888.1
|
|
Max : -1003887
|
|
|
-1003889
|
:
|
4998
|
(
|
53.3%
|
)
|
|
-1003887
|
:
|
4387
|
(
|
46.7%
|
)
|
|

|
506028 (98.2%)
|
|
ggt_ul [numeric]
|
|
Mean (sd) : 32.4 (71.5)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 18.7 ≤ 2746.8
|
|
IQR (CV) : 17.3 (2.2)
|
|
1466 distinct values
|

|
498575 (96.7%)
|
|
ggt_invalid
[factor]
|
|
|
16835
|
(
|
100.0%
|
)
|
|
3
|
(
|
0.0%
|
)
|
|
|
498575 (96.7%)
|
|
ggt_below_range
[factor]
|
|
|
16820
|
(
|
99.9%
|
)
|
|
18
|
(
|
0.1%
|
)
|
|
|
498575 (96.7%)
|
|
ggt_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result may
|
- AU680 result: {Result is
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- “Result obtained from 1 i
|
|
[ 37 others ]
|
|
|
396
|
(
|
76.0%
|
)
|
|
32
|
(
|
6.1%
|
)
|
|
23
|
(
|
4.4%
|
)
|
|
10
|
(
|
1.9%
|
)
|
|
6
|
(
|
1.2%
|
)
|
|
4
|
(
|
0.8%
|
)
|
|
3
|
(
|
0.6%
|
)
|
|
3
|
(
|
0.6%
|
)
|
|
3
|
(
|
0.6%
|
)
|
|
2
|
(
|
0.4%
|
)
|
|
39
|
(
|
7.5%
|
)
|
|

|
514892 (99.9%)
|
|
ggt_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
16838
|
(
|
100.0%
|
)
|
|

|
498575 (96.7%)
|
|
hdl_mmoll
[numeric]
|
|
Mean (sd) : 1.2 (0.3)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 1.2 ≤ 3.5
|
|
IQR (CV) : 0.4 (0.2)
|
|
236 distinct values
|
|
497230 (96.5%)
|
|
hdl_invalid
[factor]
|
|
|
18181
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
hdl_below_range
[factor]
|
|
|
18183
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
hdl_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 10 others ]
|
|
|
39
|
(
|
39.0%
|
)
|
|
28
|
(
|
28.0%
|
)
|
|
7
|
(
|
7.0%
|
)
|
|
6
|
(
|
6.0%
|
)
|
|
4
|
(
|
4.0%
|
)
|
|
2
|
(
|
2.0%
|
)
|
|
1
|
(
|
1.0%
|
)
|
|
1
|
(
|
1.0%
|
)
|
|
1
|
(
|
1.0%
|
)
|
|
1
|
(
|
1.0%
|
)
|
|
10
|
(
|
10.0%
|
)
|
|

|
515313 (100.0%)
|
|
hdl_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
18183
|
(
|
100.0%
|
)
|
|

|
497230 (96.5%)
|
|
hscrp_mgl
[numeric]
|
|
Mean (sd) : 2.5 (7.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 1 ≤ 303.3
|
|
IQR (CV) : 1.7 (2.8)
|
|
1538 distinct values
|

|
497230 (96.5%)
|
|
hscrp_invalid
[factor]
|
|
|
18180
|
(
|
100.0%
|
)
|
|
3
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
hscrp_below_range
[factor]
|
|
|
18148
|
(
|
99.8%
|
)
|
|
35
|
(
|
0.2%
|
)
|
|
|
497230 (96.5%)
|
|
hscrp_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 analysis: {HEMOL =
|
- “hsCRP result obtained fr
|
- AU680 result: {Result may
|
- AU680 result: {Result is
|
- AU680 result: {Result is
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {Result is
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
|
[ 39 others ]
|
|
|
548
|
(
|
74.5%
|
)
|
|
41
|
(
|
5.6%
|
)
|
|
31
|
(
|
4.2%
|
)
|
|
29
|
(
|
3.9%
|
)
|
|
11
|
(
|
1.5%
|
)
|
|
9
|
(
|
1.2%
|
)
|
|
7
|
(
|
1.0%
|
)
|
|
6
|
(
|
0.8%
|
)
|
|
5
|
(
|
0.7%
|
)
|
|
3
|
(
|
0.4%
|
)
|
|
46
|
(
|
6.2%
|
)
|
|

|
514677 (99.9%)
|
|
hscrp_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
18183
|
(
|
100.0%
|
)
|
|

|
497230 (96.5%)
|
|
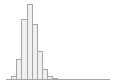
ldl_mmoll
[numeric]
|
|
Mean (sd) : 2.4 (0.7)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 2.3 ≤ 9.9
|
|
IQR (CV) : 0.9 (0.3)
|
|
484 distinct values
|
|
497230 (96.5%)
|
|
ldl_invalid
[factor]
|
|
|
18181
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
ldl_below_range
[factor]
|
|
|
18183
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
ldl_note
[character]
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 14 others ]
|
|
|
103
|
(
|
62.4%
|
)
|
|
29
|
(
|
17.6%
|
)
|
|
6
|
(
|
3.6%
|
)
|
|
4
|
(
|
2.4%
|
)
|
|
3
|
(
|
1.8%
|
)
|
|
2
|
(
|
1.2%
|
)
|
|
1
|
(
|
0.6%
|
)
|
|
1
|
(
|
0.6%
|
)
|
|
1
|
(
|
0.6%
|
)
|
|
1
|
(
|
0.6%
|
)
|
|
14
|
(
|
8.5%
|
)
|
|

|
515248 (100.0%)
|
|
ldl_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
18183
|
(
|
100.0%
|
)
|
|

|
497230 (96.5%)
|
|
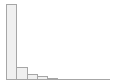
lpa_nmoll
[numeric]
|
|
Mean (sd) : 37.4 (47.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 18.5 ≤ 481.1
|
|
IQR (CV) : 35.6 (1.3)
|
|
7508 distinct values
|
|
497230 (96.5%)
|
|
lpa_invalid
[factor]
|
|
|
18179
|
(
|
100.0%
|
)
|
|
4
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
lpa_below_range
[factor]
|
|
|
17461
|
(
|
96.0%
|
)
|
|
722
|
(
|
4.0%
|
)
|
|
|
497230 (96.5%)
|
|
lpa_note
[character]
|
- AU680 result: {Result is
|
- AU680 result: {Result is
|
- AU680 analysis: {HEMOL =
|
- “Lp(a) result obtained fr
|
- AU680 result: {LIH test n
|
- AU680 analysis: {HEMOL =
|
- AU680 result: {LIH test n
|
- “Lp(a) result obtained fr
|
- AU680 result: {Result is
|
- AU680 analysis: {HEMOL =
|
|
[ 107 others ]
|
|
|
3977
|
(
|
65.9%
|
)
|
|
524
|
(
|
8.7%
|
)
|
|
423
|
(
|
7.0%
|
)
|
|
245
|
(
|
4.1%
|
)
|
|
191
|
(
|
3.2%
|
)
|
|
82
|
(
|
1.4%
|
)
|
|
79
|
(
|
1.3%
|
)
|
|
74
|
(
|
1.2%
|
)
|
|
73
|
(
|
1.2%
|
)
|
|
35
|
(
|
0.6%
|
)
|
|
333
|
(
|
5.5%
|
)
|
|

|
509377 (98.8%)
|
|
lpa_machine_id
[integer]
|
|
Min : -1019429
|
|
Mean : -1019349.6
|
|
Max : -1019349
|
|
|
-1019429
|
:
|
135
|
(
|
0.7%
|
)
|
|
-1019349
|
:
|
18048
|
(
|
99.3%
|
)
|
|

|
497230 (96.5%)
|
|
tg_mmoll [numeric]
|
|
Mean (sd) : 2 (1.6)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 1.6 ≤ 37.9
|
|
IQR (CV) : 1.3 (0.8)
|
|
843 distinct values
|
|
497230 (96.5%)
|
|
trig_invalid
[factor]
|
|
|
18181
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
trig_below_range
[factor]
|
|
|
18183
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
497230 (96.5%)
|
|
trig_note
[character]
|
- AU680 result: {LIH test n
|
- “TRIG result obtained fro
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- “Result obtained from 1 i
|
- {RES EDIT OLD=8.86}{DILUT
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 28 others ]
|
|
|
39
|
(
|
32.8%
|
)
|
|
18
|
(
|
15.1%
|
)
|
|
14
|
(
|
11.8%
|
)
|
|
7
|
(
|
5.9%
|
)
|
|
7
|
(
|
5.9%
|
)
|
|
2
|
(
|
1.7%
|
)
|
|
1
|
(
|
0.8%
|
)
|
|
1
|
(
|
0.8%
|
)
|
|
1
|
(
|
0.8%
|
)
|
|
1
|
(
|
0.8%
|
)
|
|
28
|
(
|
23.5%
|
)
|
|

|
515294 (100.0%)
|
|
trig_machine_id
[integer]
|
|
Min : -1019429
|
|
Mean : -1019349.4
|
|
Max : -1019349
|
|
|
-1019429
|
:
|
86
|
(
|
0.5%
|
)
|
|
-1019349
|
:
|
18097
|
(
|
99.5%
|
)
|
|

|
497230 (96.5%)
|
|
totalvitd_ngml
[numeric]
|
|
Mean (sd) : 33.6 (10.7)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 32 ≤ 126
|
|
IQR (CV) : 13.6 (0.3)
|
|
7980 distinct values
|
|
506290 (98.2%)
|
|
totalvitd_invalid
[factor]
|
|
|
9121
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
506290 (98.2%)
|
|
totalvitd_below_range
[factor]
|
|
|
9123
|
(
|
100.0%
|
)
|
|
0
|
(
|
0.0%
|
)
|
|
|
506290 (98.2%)
|
|
totalvitd_note
[character]
|
|
|

|
515413 (100.0%)
|
|
totalvitd_machine_id
[integer]
|
1 distinct value
|
|
-12671300
|
:
|
9123
|
(
|
100.0%
|
)
|
|
|
506290 (98.2%)
|
|
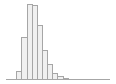
uric_umoll
[numeric]
|
|
Mean (sd) : 279.1 (83.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 269 ≤ 988
|
|
IQR (CV) : 107 (0.3)
|
|
534 distinct values
|
|
498585 (96.7%)
|
|
uric_invalid
[factor]
|
|
|
16826
|
(
|
100.0%
|
)
|
|
2
|
(
|
0.0%
|
)
|
|
|
498585 (96.7%)
|
|
uric_below_range
[factor]
|
|
|
16818
|
(
|
99.9%
|
)
|
|
10
|
(
|
0.1%
|
)
|
|

|
498585 (96.7%)
|
|
uric_note
[character]
|
- AU680 result: {Result may
|
- AU680 result: {LIH test n
|
- AU680 result: {Result may
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
- AU680 analysis: {HEMOL =
|
|
[ 21 others ]
|
|
|
27
|
(
|
29.0%
|
)
|
|
23
|
(
|
24.7%
|
)
|
|
7
|
(
|
7.5%
|
)
|
|
6
|
(
|
6.5%
|
)
|
|
4
|
(
|
4.3%
|
)
|
|
1
|
(
|
1.1%
|
)
|
|
1
|
(
|
1.1%
|
)
|
|
1
|
(
|
1.1%
|
)
|
|
1
|
(
|
1.1%
|
)
|
|
1
|
(
|
1.1%
|
)
|
|
21
|
(
|
22.6%
|
)
|
|

|
515320 (100.0%)
|
|
uric_machine_id
[integer]
|
1 distinct value
|
|
-1019349
|
:
|
16828
|
(
|
100.0%
|
)
|
|

|
498585 (96.7%)
|