|
gwas_batch
[character]
|
- A
·
|
- N
·
|
- Q
·
|
- P
·
|
- T
·
|
- J
·
|
- O
·
|
- R
·
|
- H
·
|
- C
·
|
|
[ 12 others ]
|
|
|
4685
|
(
|
4.7%
|
)
|
|
4681
|
(
|
4.6%
|
)
|
|
4679
|
(
|
4.6%
|
)
|
|
4676
|
(
|
4.6%
|
)
|
|
4669
|
(
|
4.6%
|
)
|
|
4666
|
(
|
4.6%
|
)
|
|
4663
|
(
|
4.6%
|
)
|
|
4663
|
(
|
4.6%
|
)
|
|
4661
|
(
|
4.6%
|
)
|
|
4659
|
(
|
4.6%
|
)
|
|
54004
|
(
|
53.6%
|
)
|
|

|
414707 (80.5%)
|
|
ccvid_version
[integer]
|
1 distinct value
|
|

|
414707 (80.5%)
|
|
is_in_gwas_unrelated_subset
[integer]
|
|
Min : 0
|
|
Mean : 0.8
|
|
Max : 1
|
|
|
0
|
:
|
23996
|
(
|
23.8%
|
)
|
|
1
|
:
|
76710
|
(
|
76.2%
|
)
|
|

|
414707 (80.5%)
|
|
is_in_gwas_population_subset
[integer]
|
|
Min : 0
|
|
Mean : 0.8
|
|
Max : 1
|
|
|
0
|
:
|
23530
|
(
|
23.4%
|
)
|
|
1
|
:
|
77176
|
(
|
76.6%
|
)
|
|

|
414707 (80.5%)
|
|
principal_components_source
[factor]
|
- Directly derived from par
|
- Average of sampled partic
|
|
|
100706
|
(
|
20.5%
|
)
|
|
390082
|
(
|
79.5%
|
)
|
|

|
24625 (4.8%)
|
|
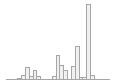
national_pc01
[numeric]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
-0.2 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0.1 (-20.2)
|
|
101321 distinct values
|
|
24625 (4.8%)
|
|
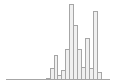
national_pc02
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (28.1)
|
|
101726 distinct values
|
|
24625 (4.8%)
|
|
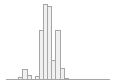
national_pc03
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (21.7)
|
|
101676 distinct values
|
|
24625 (4.8%)
|
|
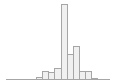
national_pc04
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (42.6)
|
|
101673 distinct values
|
|
24625 (4.8%)
|
|
national_pc05
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (-7.1)
|
|
101677 distinct values
|
|
24625 (4.8%)
|
|
national_pc06
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (-10)
|
|
101603 distinct values
|
|
24625 (4.8%)
|
|
national_pc07
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (14)
|
|
101664 distinct values
|
|
24625 (4.8%)
|
|
national_pc08
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (-9.4)
|
|
101580 distinct values
|
|
24625 (4.8%)
|
|
national_pc09
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (-22.6)
|
|
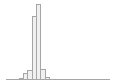
101547 distinct values
|
|
24625 (4.8%)
|
|
national_pc10
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (21.3)
|
|
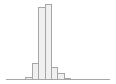
101521 distinct values
|
|
24625 (4.8%)
|
|
national_pc11
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (31.1)
|
|
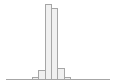
101515 distinct values
|
|
24625 (4.8%)
|
|
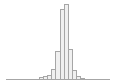
regional_pc01
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 0
|
|
IQR (CV) : 0 (-10)
|
|
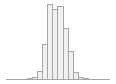
93191 distinct values
|
|
30407 (5.9%)
|
|
regional_pc02
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (-16.7)
|
|
93783 distinct values
|
|
30407 (5.9%)
|
|
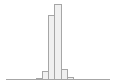
regional_pc03
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (10.8)
|
|
81705 distinct values
|
|
80154 (15.6%)
|
|
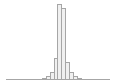
regional_pc04
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (-34.9)
|
|
74030 distinct values
|
|
105509 (20.5%)
|
|
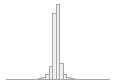
regional_pc05
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (43.3)
|
|
67630 distinct values
|
|
150566 (29.2%)
|
|
regional_pc06
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (-42.2)
|
|
51062 distinct values
|
|
236478 (45.9%)
|
|
regional_pc07
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (-41.9)
|
|
40556 distinct values
|

|
291661 (56.6%)
|
|
regional_pc08
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (12635.8)
|
|
40579 distinct values
|
|
291661 (56.6%)
|
|
regional_pc09
[numeric]
|
|
Mean (sd) : 0 (0)
|
|
min ≤ med ≤ max:
|
|
-0.1 ≤ 0 ≤ 0.1
|
|
IQR (CV) : 0 (284.8)
|
|
19395 distinct values
|
|
403733 (78.3%)
|
|
is_immigrant
[factor]
|
|
|
94924
|
(
|
94.3%
|
)
|
|
5782
|
(
|
5.7%
|
)
|
|
|
414707 (80.5%)
|
|
gwas_array_type
[character]
|
|
|
32265
|
(
|
32.0%
|
)
|
|
68441
|
(
|
68.0%
|
)
|
|

|
414707 (80.5%)
|