|
cimt_staff_id
[character]
|
- 161
|
- 145
|
- 162
|
- 158
|
- 148
|
- 144
|
- 151
|
- 159
|
- 149
|
- 184
|
|
[ 16 others ]
|
|
|
3077
|
(
|
12.2%
|
)
|
|
2940
|
(
|
11.7%
|
)
|
|
2914
|
(
|
11.6%
|
)
|
|
2849
|
(
|
11.3%
|
)
|
|
2793
|
(
|
11.1%
|
)
|
|
2215
|
(
|
8.8%
|
)
|
|
2051
|
(
|
8.2%
|
)
|
|
1520
|
(
|
6.0%
|
)
|
|
1393
|
(
|
5.5%
|
)
|
|
1362
|
(
|
5.4%
|
)
|
|
2032
|
(
|
8.1%
|
)
|
|

|
106 (0.4%)
|
|
cimt_race
[character]
|
|
|
8899
|
(
|
35.4%
|
)
|
|
16247
|
(
|
64.6%
|
)
|
|

|
106 (0.4%)
|
|
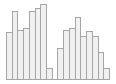
cimt_resultsdateandtime
[POSIXct, POSIXt]
|
|
min : 2013-08-04 00:49:07
|
|
med : 2013-12-31 01:58:58
|
|
max : 2014-09-18 01:53:17
|
|
range : 1y 1m 14d 1H 4M 10S
|
|
25121 distinct values
|
|
106 (0.4%)
|
|
cimt_rplq
[character]
|
|
|
17679
|
(
|
70.3%
|
)
|
|
7463
|
(
|
29.7%
|
)
|
|

|
110 (0.4%)
|
|
cimt_lplq
[character]
|
|
|
17395
|
(
|
69.2%
|
)
|
|
7745
|
(
|
30.8%
|
)
|
|

|
112 (0.4%)
|
|
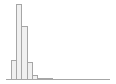
cimt_r_maximt_report
[numeric]
|
|
Mean (sd) : 0.8 (0.2)
|
|
min ≤ med ≤ max:
|
|
0.4 ≤ 0.8 ≤ 4.1
|
|
IQR (CV) : 0.2 (0.3)
|
|
181 distinct values
|
|
110 (0.4%)
|
|
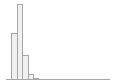
cimt_r_meanimt_report
[numeric]
|
|
Mean (sd) : 0.7 (0.2)
|
|
min ≤ med ≤ max:
|
|
0.3 ≤ 0.7 ≤ 3.9
|
|
IQR (CV) : 0.2 (0.3)
|
|
1081 distinct values
|
|
110 (0.4%)
|
|
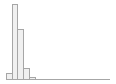
cimt_r_minimt_report
[numeric]
|
|
Mean (sd) : 0.6 (0.2)
|
|
min ≤ med ≤ max:
|
|
0.3 ≤ 0.6 ≤ 3.7
|
|
IQR (CV) : 0.2 (0.3)
|
|
141 distinct values
|
|
110 (0.4%)
|
|
cimt_r_imt_ang_report
[character]
|
|
|
13644
|
(
|
54.3%
|
)
|
|
11498
|
(
|
45.7%
|
)
|
|

|
110 (0.4%)
|
|
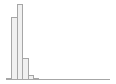
cimt_r_mean_imt_120
[numeric]
|
|
Mean (sd) : 0.7 (0.2)
|
|
min ≤ med ≤ max:
|
|
0.3 ≤ 0.6 ≤ 3.9
|
|
IQR (CV) : 0.2 (0.3)
|
|
1059 distinct values
|
|
163 (0.6%)
|
|
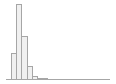
cimt_r_max_imt_120
[numeric]
|
|
Mean (sd) : 0.8 (0.2)
|
|
min ≤ med ≤ max:
|
|
0.3 ≤ 0.7 ≤ 4.1
|
|
IQR (CV) : 0.2 (0.3)
|
|
171 distinct values
|
|
163 (0.6%)
|
|
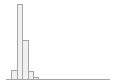
cimt_r_min_imt_120
[numeric]
|
|
Mean (sd) : 0.6 (0.2)
|
|
min ≤ med ≤ max:
|
|
0.1 ≤ 0.5 ≤ 3.7
|
|
IQR (CV) : 0.2 (0.3)
|
|
134 distinct values
|
|
163 (0.6%)
|
|
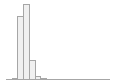
cimt_r_mean_imt_150
[numeric]
|
|
Mean (sd) : 0.7 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.6 ≤ 3.4
|
|
IQR (CV) : 0.2 (0.2)
|
|
1014 distinct values
|
|
118 (0.5%)
|
|
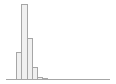
cimt_r_max_imt_150
[numeric]
|
|
Mean (sd) : 0.8 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.7 ≤ 3.9
|
|
IQR (CV) : 0.2 (0.3)
|
|
168 distinct values
|
|
118 (0.5%)
|
|
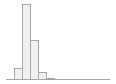
cimt_r_min_imt_150
[numeric]
|
|
Mean (sd) : 0.6 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.5 ≤ 2.5
|
|
IQR (CV) : 0.2 (0.3)
|
|
136 distinct values
|
|
118 (0.5%)
|
|
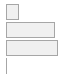
cimt_r_imt_percentile
[character]
|
-
·
<= 25%-tile
|
-
·
= 75%-tile
|
- 25 %-tile<= XX <==75 %-ti
|
- NA
|
|
|
2766
|
(
|
11.0%
|
)
|
|
10768
|
(
|
42.8%
|
)
|
|
11555
|
(
|
46.0%
|
)
|
|
56
|
(
|
0.2%
|
)
|
|
|
107 (0.4%)
|
|
cimt_rplq_count
[integer]
|
|
Mean (sd) : 0.4 (0.7)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 8
|
|
IQR (CV) : 1 (1.8)
|
|
|
0
|
:
|
17689
|
(
|
70.4%
|
)
|
|
1
|
:
|
5433
|
(
|
21.6%
|
)
|
|
2
|
:
|
1591
|
(
|
6.3%
|
)
|
|
3
|
:
|
308
|
(
|
1.2%
|
)
|
|
4
|
:
|
77
|
(
|
0.3%
|
)
|
|
5
|
:
|
25
|
(
|
0.1%
|
)
|
|
6
|
:
|
13
|
(
|
0.1%
|
)
|
|
7
|
:
|
2
|
(
|
0.0%
|
)
|
|
8
|
:
|
2
|
(
|
0.0%
|
)
|
|

|
112 (0.4%)
|
|
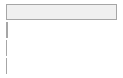
cimt_rplq_count_ica
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 3
|
|
IQR (CV) : 0 (9.5)
|
|
|
0
|
:
|
24845
|
(
|
98.8%
|
)
|
|
1
|
:
|
276
|
(
|
1.1%
|
)
|
|
2
|
:
|
18
|
(
|
0.1%
|
)
|
|
3
|
:
|
1
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
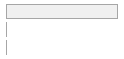
cimt_rplq_count_eca
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 2
|
|
IQR (CV) : 0 (14.2)
|
|
|
0
|
:
|
25012
|
(
|
99.5%
|
)
|
|
1
|
:
|
123
|
(
|
0.5%
|
)
|
|
2
|
:
|
5
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
cimt_rplq_count_bif
[integer]
|
|
Mean (sd) : 0.2 (0.5)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 3
|
|
IQR (CV) : 0 (2.1)
|
|
|
0
|
:
|
20023
|
(
|
79.6%
|
)
|
|
1
|
:
|
4180
|
(
|
16.6%
|
)
|
|
2
|
:
|
889
|
(
|
3.5%
|
)
|
|
3
|
:
|
48
|
(
|
0.2%
|
)
|
|
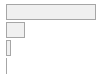
|
112 (0.4%)
|
|
cimt_rplq_count_cca
[integer]
|
|
Mean (sd) : 0.1 (0.4)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 7
|
|
IQR (CV) : 0 (3.1)
|
|
|
0
|
:
|
22254
|
(
|
88.5%
|
)
|
|
1
|
:
|
2348
|
(
|
9.3%
|
)
|
|
2
|
:
|
468
|
(
|
1.9%
|
)
|
|
3
|
:
|
45
|
(
|
0.2%
|
)
|
|
4
|
:
|
18
|
(
|
0.1%
|
)
|
|
5
|
:
|
4
|
(
|
0.0%
|
)
|
|
6
|
:
|
2
|
(
|
0.0%
|
)
|
|
7
|
:
|
1
|
(
|
0.0%
|
)
|
|

|
112 (0.4%)
|
|
cimt_rplq_distance
[numeric]
|
|
Mean (sd) : 2 (0.7)
|
|
min ≤ med ≤ max:
|
|
0.7 ≤ 1.8 ≤ 8.1
|
|
IQR (CV) : 0.9 (0.4)
|
|
1001 distinct values
|
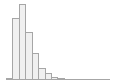
|
17787 (70.4%)
|
|
cimt_rplq_stenosis
[integer]
|
|
Mean (sd) : 4699 (9373.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 100019
|
|
IQR (CV) : 6893 (2)
|
|
6370 distinct values
|

|
112 (0.4%)
|
|
cimt_rplq_minpixelsize
[integer]
|
|
Mean (sd) : 164.3 (356.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 5628
|
|
IQR (CV) : 179 (2.2)
|
|
1612 distinct values
|
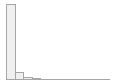
|
112 (0.4%)
|
|
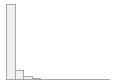
cimt_rplq_maxpixelsize
[integer]
|
|
Mean (sd) : 200 (417.8)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 5628
|
|
IQR (CV) : 241 (2.1)
|
|
1798 distinct values
|
|
112 (0.4%)
|
|
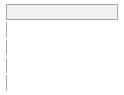
cimt_rplq_excluded_count
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 4
|
|
IQR (CV) : 0 (33.9)
|
|
|
0
|
:
|
25112
|
(
|
99.9%
|
)
|
|
1
|
:
|
21
|
(
|
0.1%
|
)
|
|
2
|
:
|
5
|
(
|
0.0%
|
)
|
|
3
|
:
|
1
|
(
|
0.0%
|
)
|
|
4
|
:
|
1
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
cimt_rplq_combined_count
[integer]
|
|
Mean (sd) : 0 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 6
|
|
IQR (CV) : 0 (8.3)
|
|
|
0
|
:
|
24703
|
(
|
98.3%
|
)
|
|
1
|
:
|
391
|
(
|
1.6%
|
)
|
|
2
|
:
|
35
|
(
|
0.1%
|
)
|
|
3
|
:
|
7
|
(
|
0.0%
|
)
|
|
4
|
:
|
1
|
(
|
0.0%
|
)
|
|
5
|
:
|
2
|
(
|
0.0%
|
)
|
|
6
|
:
|
1
|
(
|
0.0%
|
)
|
|

|
112 (0.4%)
|
|
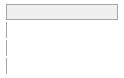
cimt_rplq_divided_count
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 3
|
|
IQR (CV) : 0 (16.3)
|
|
|
0
|
:
|
25036
|
(
|
99.6%
|
)
|
|
1
|
:
|
92
|
(
|
0.4%
|
)
|
|
2
|
:
|
11
|
(
|
0.0%
|
)
|
|
3
|
:
|
1
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
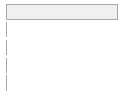
cimt_rplq_special_count
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 4
|
|
IQR (CV) : 0 (25.3)
|
|
|
0
|
:
|
25091
|
(
|
99.8%
|
)
|
|
1
|
:
|
42
|
(
|
0.2%
|
)
|
|
2
|
:
|
4
|
(
|
0.0%
|
)
|
|
3
|
:
|
2
|
(
|
0.0%
|
)
|
|
4
|
:
|
1
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
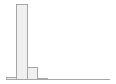
cimt_l_maximt_report
[numeric]
|
|
Mean (sd) : 0.8 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.8 ≤ 4.7
|
|
IQR (CV) : 0.2 (0.3)
|
|
184 distinct values
|
|
109 (0.4%)
|
|
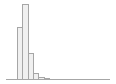
cimt_l_meanimt_report
[numeric]
|
|
Mean (sd) : 0.7 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.7 ≤ 3.6
|
|
IQR (CV) : 0.2 (0.3)
|
|
1132 distinct values
|
|
109 (0.4%)
|
|
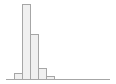
cimt_l_minimt_report
[numeric]
|
|
Mean (sd) : 0.6 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.6 ≤ 2.5
|
|
IQR (CV) : 0.2 (0.3)
|
|
146 distinct values
|
|
109 (0.4%)
|
|
cimt_l_imt_ang_report
[character]
|
|
|
11899
|
(
|
47.3%
|
)
|
|
13244
|
(
|
52.7%
|
)
|
|

|
109 (0.4%)
|
|
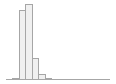
cimt_l_mean_imt_210
[numeric]
|
|
Mean (sd) : 0.7 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.6 ≤ 3.1
|
|
IQR (CV) : 0.2 (0.3)
|
|
1066 distinct values
|
|
119 (0.5%)
|
|
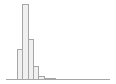
cimt_l_max_imt_210
[numeric]
|
|
Mean (sd) : 0.8 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.7 ≤ 3.8
|
|
IQR (CV) : 0.2 (0.3)
|
|
168 distinct values
|
|
119 (0.5%)
|
|
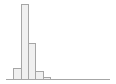
cimt_l_min_imt_210
[numeric]
|
|
Mean (sd) : 0.6 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.5 ≤ 2.8
|
|
IQR (CV) : 0.2 (0.3)
|
|
136 distinct values
|
|
119 (0.5%)
|
|
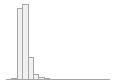
cimt_l_mean_imt_240
[numeric]
|
|
Mean (sd) : 0.7 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.6 ≤ 3.6
|
|
IQR (CV) : 0.2 (0.3)
|
|
1096 distinct values
|
|
241 (1.0%)
|
|
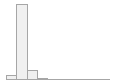
cimt_l_max_imt_240
[numeric]
|
|
Mean (sd) : 0.8 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.7 ≤ 4.7
|
|
IQR (CV) : 0.2 (0.3)
|
|
178 distinct values
|
|
241 (1.0%)
|
|
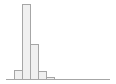
cimt_l_min_imt_240
[numeric]
|
|
Mean (sd) : 0.6 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0.5 ≤ 2.5
|
|
IQR (CV) : 0.2 (0.3)
|
|
146 distinct values
|
|
241 (1.0%)
|
|
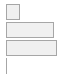
cimt_l_imt_percentile
[character]
|
-
·
<= 25%-tile
|
-
·
= 75%-tile
|
- 25 %-tile<= XX <==75 %-ti
|
- NA
|
|
|
3021
|
(
|
12.0%
|
)
|
|
10678
|
(
|
42.5%
|
)
|
|
11391
|
(
|
45.3%
|
)
|
|
55
|
(
|
0.2%
|
)
|
|
|
107 (0.4%)
|
|
cimt_lplq_count
[integer]
|
|
Mean (sd) : 0.4 (0.7)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 8
|
|
IQR (CV) : 1 (1.8)
|
|
|
0
|
:
|
17424
|
(
|
69.3%
|
)
|
|
1
|
:
|
5550
|
(
|
22.1%
|
)
|
|
2
|
:
|
1655
|
(
|
6.6%
|
)
|
|
3
|
:
|
357
|
(
|
1.4%
|
)
|
|
4
|
:
|
95
|
(
|
0.4%
|
)
|
|
5
|
:
|
43
|
(
|
0.2%
|
)
|
|
6
|
:
|
13
|
(
|
0.1%
|
)
|
|
7
|
:
|
1
|
(
|
0.0%
|
)
|
|
8
|
:
|
2
|
(
|
0.0%
|
)
|
|

|
112 (0.4%)
|
|
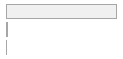
cimt_lplq_count_ica
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 2
|
|
IQR (CV) : 0 (9.7)
|
|
|
0
|
:
|
24863
|
(
|
98.9%
|
)
|
|
1
|
:
|
259
|
(
|
1.0%
|
)
|
|
2
|
:
|
18
|
(
|
0.1%
|
)
|
|
|
112 (0.4%)
|
|
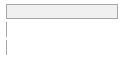
cimt_lplq_count_eca
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 2
|
|
IQR (CV) : 0 (13.5)
|
|
|
0
|
:
|
24998
|
(
|
99.4%
|
)
|
|
1
|
:
|
137
|
(
|
0.5%
|
)
|
|
2
|
:
|
5
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
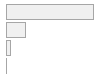
cimt_lplq_count_bif
[integer]
|
|
Mean (sd) : 0.3 (0.5)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 3
|
|
IQR (CV) : 0 (2.1)
|
|
|
0
|
:
|
19763
|
(
|
78.6%
|
)
|
|
1
|
:
|
4337
|
(
|
17.3%
|
)
|
|
2
|
:
|
985
|
(
|
3.9%
|
)
|
|
3
|
:
|
55
|
(
|
0.2%
|
)
|
|
|
112 (0.4%)
|
|
cimt_lplq_count_cca
[integer]
|
|
Mean (sd) : 0.1 (0.4)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 7
|
|
IQR (CV) : 0 (3)
|
|
|
0
|
:
|
22127
|
(
|
88.0%
|
)
|
|
1
|
:
|
2424
|
(
|
9.6%
|
)
|
|
2
|
:
|
505
|
(
|
2.0%
|
)
|
|
3
|
:
|
64
|
(
|
0.3%
|
)
|
|
4
|
:
|
16
|
(
|
0.1%
|
)
|
|
5
|
:
|
2
|
(
|
0.0%
|
)
|
|
6
|
:
|
1
|
(
|
0.0%
|
)
|
|
7
|
:
|
1
|
(
|
0.0%
|
)
|
|

|
112 (0.4%)
|
|
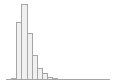
cimt_lplq_distance
[numeric]
|
|
Mean (sd) : 2 (0.7)
|
|
min ≤ med ≤ max:
|
|
0.4 ≤ 1.8 ≤ 9.9
|
|
IQR (CV) : 0.8 (0.4)
|
|
1004 distinct values
|
|
17517 (69.4%)
|
|
cimt_lplq_stenosis
[integer]
|
|
Mean (sd) : 4926.3 (9592)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 100027
|
|
IQR (CV) : 7453.2 (1.9)
|
|
6605 distinct values
|

|
112 (0.4%)
|
|
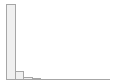
cimt_lplq_minpixelsize
[integer]
|
|
Mean (sd) : 171.7 (367)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 5998
|
|
IQR (CV) : 197 (2.1)
|
|
1642 distinct values
|
|
112 (0.4%)
|
|
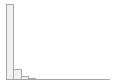
cimt_lplq_maxpixelsize
[integer]
|
|
Mean (sd) : 211.9 (437.3)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 6610
|
|
IQR (CV) : 276 (2.1)
|
|
1823 distinct values
|
|
112 (0.4%)
|
|
cimt_lplq_excluded_count
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 5
|
|
IQR (CV) : 0 (29.6)
|
|
|
0
|
:
|
25102
|
(
|
99.8%
|
)
|
|
1
|
:
|
33
|
(
|
0.1%
|
)
|
|
2
|
:
|
4
|
(
|
0.0%
|
)
|
|
5
|
:
|
1
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
cimt_lplq_combined_count
[integer]
|
|
Mean (sd) : 0 (0.2)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 8
|
|
IQR (CV) : 0 (7.9)
|
|
|
0
|
:
|
24640
|
(
|
98.0%
|
)
|
|
1
|
:
|
446
|
(
|
1.8%
|
)
|
|
2
|
:
|
38
|
(
|
0.2%
|
)
|
|
3
|
:
|
10
|
(
|
0.0%
|
)
|
|
4
|
:
|
4
|
(
|
0.0%
|
)
|
|
6
|
:
|
1
|
(
|
0.0%
|
)
|
|
8
|
:
|
1
|
(
|
0.0%
|
)
|
|

|
112 (0.4%)
|
|
cimt_lplq_divided_count
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 3
|
|
IQR (CV) : 0 (15.1)
|
|
|
0
|
:
|
25016
|
(
|
99.5%
|
)
|
|
1
|
:
|
117
|
(
|
0.5%
|
)
|
|
2
|
:
|
3
|
(
|
0.0%
|
)
|
|
3
|
:
|
4
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
cimt_lplq_special_count
[integer]
|
|
Mean (sd) : 0 (0.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 4
|
|
IQR (CV) : 0 (25.9)
|
|
|
0
|
:
|
25095
|
(
|
99.8%
|
)
|
|
1
|
:
|
40
|
(
|
0.2%
|
)
|
|
2
|
:
|
4
|
(
|
0.0%
|
)
|
|
4
|
:
|
1
|
(
|
0.0%
|
)
|
|
|
112 (0.4%)
|
|
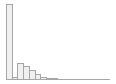
cimt_plq_score
[numeric]
|
|
Mean (sd) : 0.8 (1.1)
|
|
min ≤ med ≤ max:
|
|
0 ≤ 0 ≤ 8.7
|
|
IQR (CV) : 1.5 (1.4)
|
|
3037 distinct values
|
|
113 (0.4%)
|
|
cimt_device_manufacturer
[character]
|
- Panasonic
|
|

|
106 (0.4%)
|
|
cimt_device_name
[character]
|
- CardioHealth Station
|
|

|
106 (0.4%)
|
|
cimt_deviceid
[character]
|
- A0U2X0119
|
- A0U2X0123
|
- A0U2X0124
|
|
|
10991
|
(
|
43.7%
|
)
|
|
8899
|
(
|
35.4%
|
)
|
|
5256
|
(
|
20.9%
|
)
|
|

|
106 (0.4%)
|




























































